1. What is PendoTMBase ?
PeTMbase is a specialized web-based database platform to provide detailed information of plant eTMs. We have presentd a total of 2669 nonredundancy eTM records in 43 plant species. The platform also provides a function of predict novel eTMs based on miRNA sequences submitted by users.The number of deposited eTM records are being increased as the new species are dissected for target mimicry and all eTM sequences are available for download.
2.What is the miRNA format ?
A sequence in FASTA format begins with a single-line description, followed by lines of sequence data. At the beginning, the description line is distinguished from the sequence data by ">" symbol and includs species, miRNA' name and other informations about this miRNA . It is recommended that all lines of text be shorter than 50 characters in length. An example sequence in FASTA format is:

3.What is the eTM model format ?
Based on the model of the experiment eTM sequence, we insert a 3nt fragment of bulge between the 9th and 12th base of the miRNA sequence in the direction of 5' to 3'.

4.How to search ?
You can search by miRNA name e.g. “miR159” or “Bdi-miR827- 3p”, or select a species name to list all available eTMs regarding the species of interest. The below shows how to search and to see results in PetMbase database.
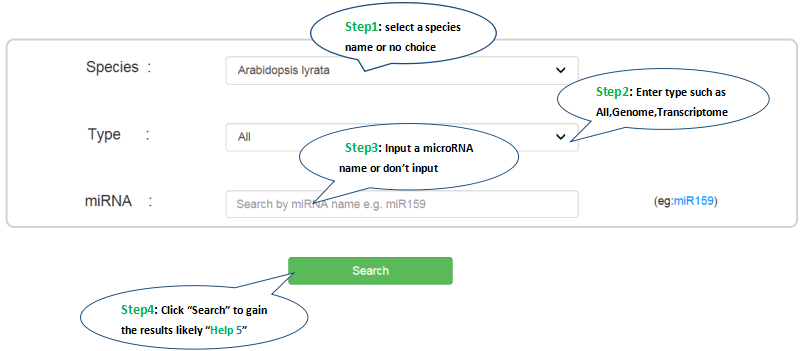
5.What is the eTM result ?
The result page(s) will present the list of searched eTMs and some important information for each query conditions. More detailed information of eTM can be found by clicking on the filename.Then we can see the follows:
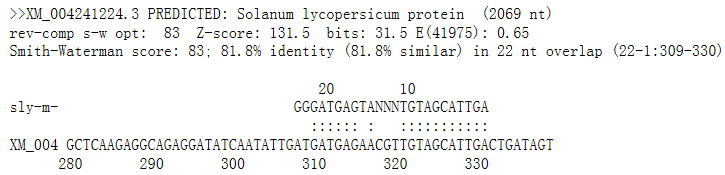
6.How can I download eTMs ?
You can freely download all available eTMs sequence and binding site information in per species. Please select and click the corresponding link of each species in the download section. The data in this database are freely available for all academic and non-commercial users. Commercial users are kindly requested to contact us for details of possible licensing arrangements.
